Curve fitting with genetic algorithm
Before reading this blog it would be great if you could read and get familiarized with Guessing a Number with Genetic Algorithm.
In this blog we would like to fit a line with genetic algorithm for the equation \(y = 7x + 5\). So let’s write the equation in Julia
y(x) = 7x + 5
Output:
y (generic function with 1 method)
Now let’s assign x to range 1 to 5 in steps of 0.1
x = 0 : 0.1 : 5
Output:
0.0:0.1:5.0
Now let’s plot the line
using Plots
scatter_plot = scatter(x, y.(x), xlims = (-0.1, 5.5), ylims = (0, 50))
There is a property in Julia which I want you to remember, let’s say we have a tuple (5, 7) and we assign it to two variables m and c as shown below
m, c = (5, 7)
println("m = ", m)
println("c = ", c)
Output:
m = 5
c = 7
Then the first variable will take the first value of the tuple and second will take the second value. We will be using this technique in this blog.
For genetic algorithm, mutation or change is very important. Let’s write a mutation function for a two valued tuple as shown:
function mutate(value, mutations = 10)
[value + rand() - 0.5 for i in 1:mutations]
end
function mutate(mc::Tuple, number_of_mutations = 10)
m, c = mc
ms = mutate(m, number_of_mutations)
cs = mutate(c, number_of_mutations)
[(ms[i], cs[i]) for i in 1:number_of_mutations]
end
Output:
mutate (generic function with 4 methods)
and we test that function:
mutate((1, 2))
10-element Vector{Tuple{Float64, Float64}}:
(1.1205168123693385, 2.41514037266611)
(0.5079882676843603, 1.8217999723149112)
(1.1581739571074665, 2.459460436379917)
(0.5796738025677814, 1.9702636743774655)
(1.0321606616260794, 1.7632954510809746)
(1.3545156402167784, 1.8782505949943635)
(1.3407759822406362, 2.3597713445300306)
(0.8524954649600578, 2.4966166282430047)
(0.991673117157789, 2.4749204653332812)
(0.9933905544923662, 1.5623361787716572)
Now let me explain how it works. First we have a mutation function that takes single value and mutates it as shown below, to learn it you may go to this blog.
function mutate(value, mutations = 10)
[value + rand() - 0.5 for i in 1:mutations]
end
Now we kind of want to extend this mutate function to tuple, so we write a declaration for it as shown
function mutate(value, mutations = 10)
[value + rand() - 0.5 for i in 1:mutations]
end
function mutate(mc::Tuple, number_of_mutations = 10)
end
Note that unlike the mutate(value, mutations = 10) this mutate(mc::Tuple, number_of_mutations = 10) clearly says it needs a tuple. Now imagine I pass a tuple to mutate like mutate((1, 2)), clearly Julia will pass it to mutate(mc::Tuple, number_of_mutations = 10) and not to mutate(value, mutations = 10). So now let’s define the tuple mutate.
Let’s assume the tuple will have only two values, so the first value should be assigned to a variable m and second one to c as shown
function mutate(mc::Tuple, number_of_mutations = 10)
m, c = mc
end
Now all we need to do is to mutate m and c by number_of_mutations times and combine it. To mutate m and c we use the following code
ms = mutate(m, number_of_mutations)
cs = mutate(c, number_of_mutations)
In the above code mutate(m, number_of_mutations) and mutate(c, number_of_mutations) calls mutate(value, mutations = 10). We store the values of mutation in variable ms, I pronounce it as emmmss and other in seas cs, let’s plugin this code into our function as shown
function mutate(mc::Tuple, number_of_mutations = 10)
m, c = mc
ms = mutate(m, number_of_mutations)
cs = mutate(c, number_of_mutations)
end
Now we combine these mutations to form a Array of Tuple’s and return it as shown
function mutate(mc::Tuple, number_of_mutations = 10)
m, c = mc
ms = mutate(m, number_of_mutations)
cs = mutate(c, number_of_mutations)
[(ms[i], cs[i]) for i in 1:number_of_mutations]
end
Now let’s write an error function, let’s say that we have m and c that our algorithm recommends, for a given x, the prediction will be (m * x + c) , and in the training phase the value of y may vary from our prediction, so, we need to minus y from our prediction to get error as shown:
∆(m, c, x, y) = (m * x + c) - y
Output:
∆ (generic function with 1 method)
Let’s say our m is 5, and c is 4, let’s see what’s out error when x is 10 as shown below
∆(5, 4, 10, y(10))
Output:
-21
Now some one says he has 10 values of \(x\) and corresponding \(y\) with him, he wants us to fit a line to predict the unknown stuff, we decide a value of \(m\) and \(c\), we need to know the total error for all values of our prediction, so let’s write a total_error function as shown:
function total_error(m, c, x, y)
ΣΔ = 0
for i in 1:length(x)
ΣΔ += abs(∆(m, c, x[i], y[i]))
end
ΣΔ
end
output:
total_error (generic function with 1 method)
In the total_error, we have a variable ΣΔ to store the total error, we assign it to zero as shown:
function total_error(m, c, x, y)
ΣΔ = 0
end
now for each element if x
function total_error(m, c, x, y)
ΣΔ = 0
for i in 1:length(x)
end
end
we add absolute value of error to total error variable ΣΔ
function total_error(m, c, x, y)
ΣΔ = 0
for i in 1:length(x)
ΣΔ += abs(∆(m, c, x[i], y[i]))
end
end
we return the total error variable ΣΔ
function total_error(m, c, x, y)
ΣΔ = 0
for i in 1:length(x)
ΣΔ += abs(∆(m, c, x[i], y[i]))
end
ΣΔ
end
Now let’s check the total_error function
total_error(5, 4, [1, 2, 3, 4, 5], [1, 2, 3, 4, 5])
Output:
80
Seems to work!
Let’s now write our top_survivors function, this one takes Array of \(m\)’s and \(c\)’s Tuple’s as mcs (I spell it as emm seee’s), an array of training inputs namely x_train and y_train, and selects the top_percent of mcs that that deviates the least with y_train for the given x_train.
function top_survivors(mcs, x_train, y_train, top_percent = 10)
errors_and_values = []
for mc in mcs
m, c = mc
error = total_error(m, c, x_train, y_train)
push!(errors_and_values, (error, mc))
end
sorted_errors_and_values = sort(errors_and_values)
end_number = Int(length(mcs) * top_percent / 100)
sorted_errors_and_values[1:end_number]
end
Output:
top_survivors (generic function with 2 methods)
I would encourage you to read my previous Genetic Algorithm blog to understand top_survivors function.
Now let’s setup the needed inputs for top_survivors, let’s have initial mcs as (0, 0):
x_train = [1, 2, 3, 4, 5]
y_train = y.(x_train)
mcs = mutate((0, 0))
Output:
10-element Vector{Tuple{Float64, Float64}}:
(0.16087862232389538, 0.018092305432591882)
(0.08312155751992667, -0.022630590705241538)
(-0.32885966826028445, -0.4936484604469795)
(-0.19041016854936288, 0.4707757045419352)
(0.011998626752193209, -0.276041600093212)
(-0.18351337891221542, -0.31274629114220875)
(0.014823442262999142, 0.03761731346306618)
(-0.19617757028312166, -0.11227454061420317)
(0.07142332361410042, -0.2765731664220268)
(0.3720719485797561, 0.44332686459739223)
Let’s test it out:
top_survivors(mcs, x_train, y_train)
Output:
1-element Vector{Any}:
(122.2022864483167, (0.3720719485797561, 0.44332686459739223))
Looks like our m of 0.3720719485797561 and c of 0.44332686459739223 are the fittest with an error of 122.2022864483167. Now all we need to do it to tell it to select top survivors across many generations so that we get a near perfect fit as shown:
generations = 40
top_survivor = Nothing
mc = (0, 0)
for i in 1:generations
mcs = mutate(mc)
top_survivor = top_survivors(mcs, x_train, y_train)[1]
_error, mc = top_survivor
end
hm, hc = mc
p = scatter(x_train, y_train)
h(x) = hm * x + hc
plot!(p, x, h.(x))
Output:
The code below is almost the same as code above, but note the @gif which collects the plot images into a gif file so that we can see it as animation:
generations = 40
top_survivor = Nothing
mc = (0, 0)
@gif for i in 1:generations
mcs = mutate(mc)
top_survivor = top_survivors(mcs, x_train, y_train)[1]
_error, mc = top_survivor
hm, hc = mc
p = scatter(x_train, y_train)
h(x) = hm * x + hc
plot!(p, x, h.(x), ylims = (0, 50))
end
Output:
┌ Info: Saved animation to
│ fn = /Users/mindaslab/data_science_with_julia/code/tmp.gif
└ @ Plots /Users/mindaslab/.julia/packages/Plots/g581z/src/animation.jl:104
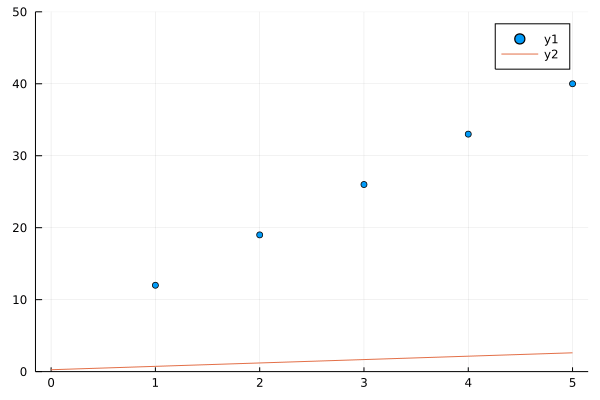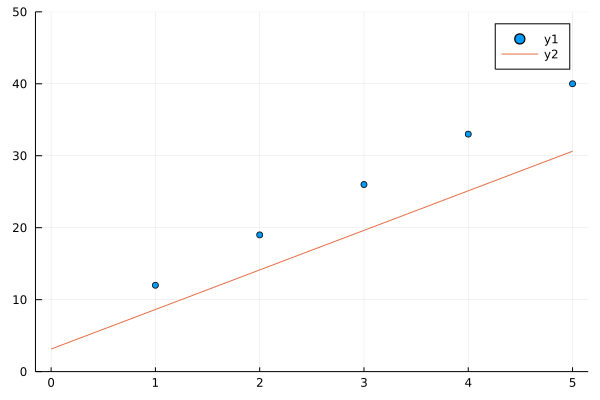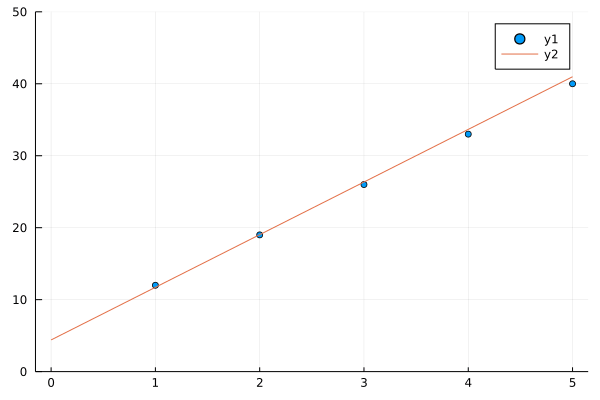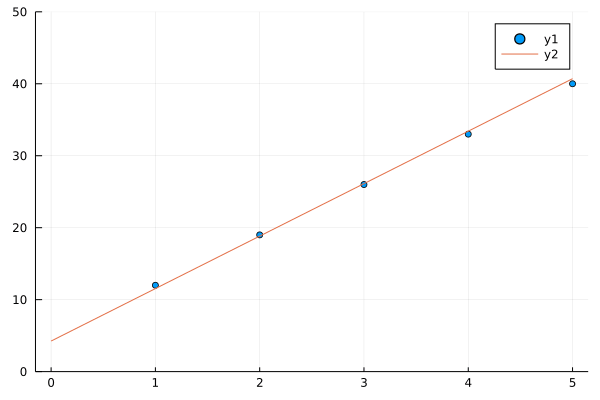
See how in 40 generations our algorithms nearly fits a line to y(x) = 7x + 5, you may change 7 and 5 in y(x) to any thing and our algorithm in successive generations will try to fit line to a bunch of points better and better in successive generations.
When I tried, this was the final value I got for \(m\) and $$c$
mc
Output:
(6.838106002778459, 5.334256635688782)
In your computer it could vary, but the line will be a near perfect fit for the bunch of points.
You can get the code for this blog here https://gitlab.com/data-science-with-julia/code/-/blob/master/genetic_alogorithms_line_fitting.ipynb.

